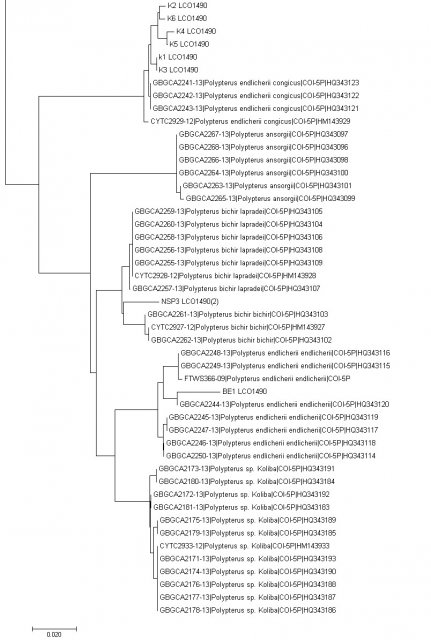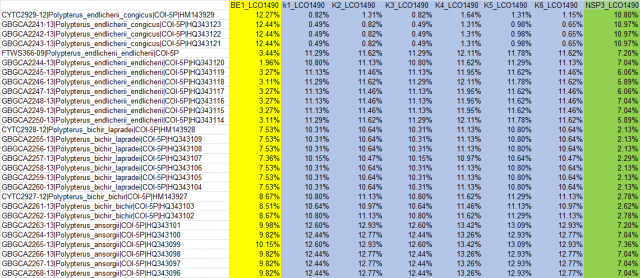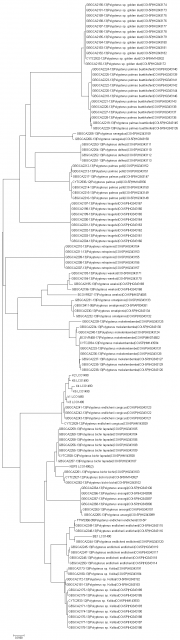
Neighbor joining tree of extant Polypterus species with Mitocondrial cytochrome C cooxydase subunit 1 (mtCOI) i made lately for graduation paper.
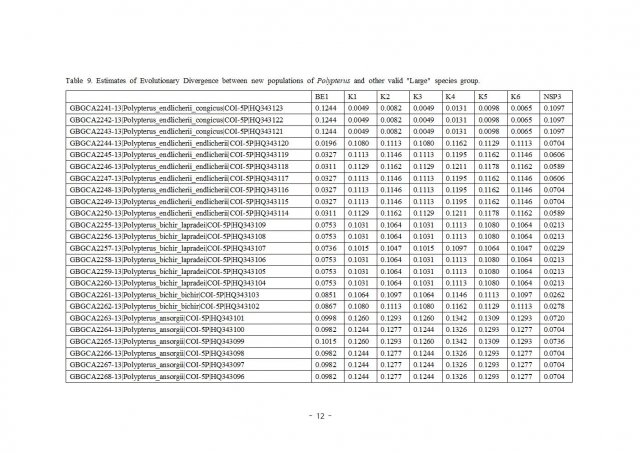
K1~K6 are stands for Giselas bichir that catched from Kirando area, East coast of Lake Tanganyika.
BE1 is stand for Polypterus endlicheri from Black volta river.
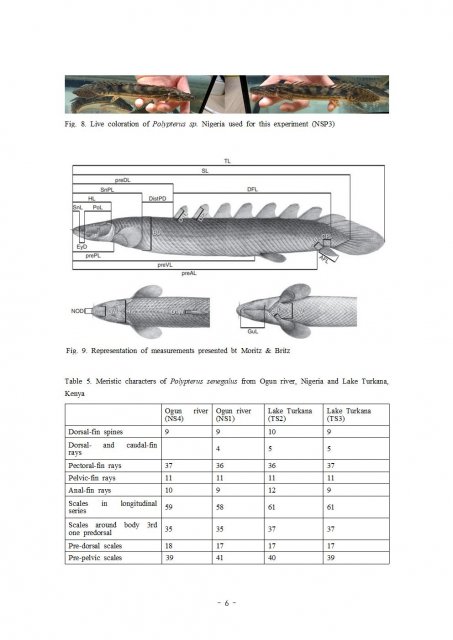
NSP3 is stands for Polypterus sp. "Nigeria" formally i posted as Polypterus sp. aff. "bichir".
It's too small number of samples to make sure these are different species or not but still very interesting result for sure.
Rest of samples are from Boldsystems and many of them are quite an old data. (that's why some of them has non-valid name now)
And also i still don't have no idea what is "sp.Koliba" on that sequence sample. (It might can be just endlicheri from Koliba area or something else because i can't find any photo of the sample)
If i get any chance to get more numbers of samples, i'll try this again with more samples with other gene like 16srRNA too.
I don't know that i will write my paper on English or Korean yet but i will share if i write it on English.
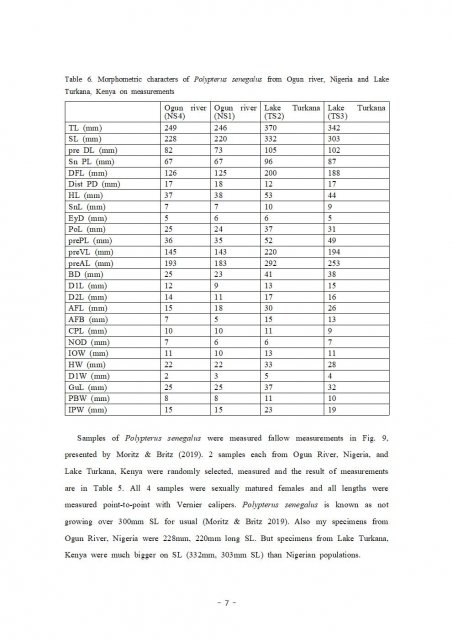
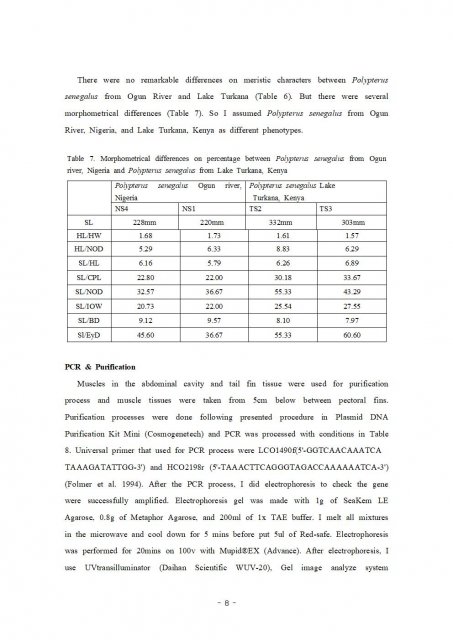
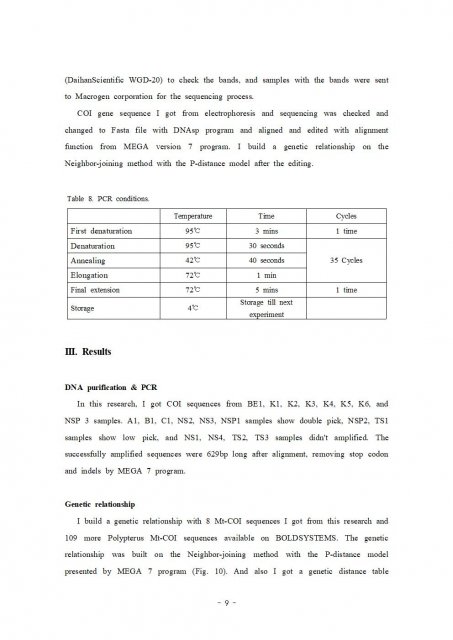
This tree made with MEGA 7 program.